Latest News from the NCDIR
Research Spotlight – NCDIR scientists have developed ODELAM, a technique to study drug resistance in Mycobacterium tuberculosis
NCDIR scientists led by Dr. Thurston Herricks in the lab of Prof. John Aitchison (Seattle Children’s) developed a novel time-lapse microcopy approach for high throughput analysis of Mycobacterium tuberculosis growth to understand its genetic-environment interactions. One very important application of this work is investigating how pathogens like Mycobacterium tuberculosis (Mtb) respond to antibiotics and develop resistance.
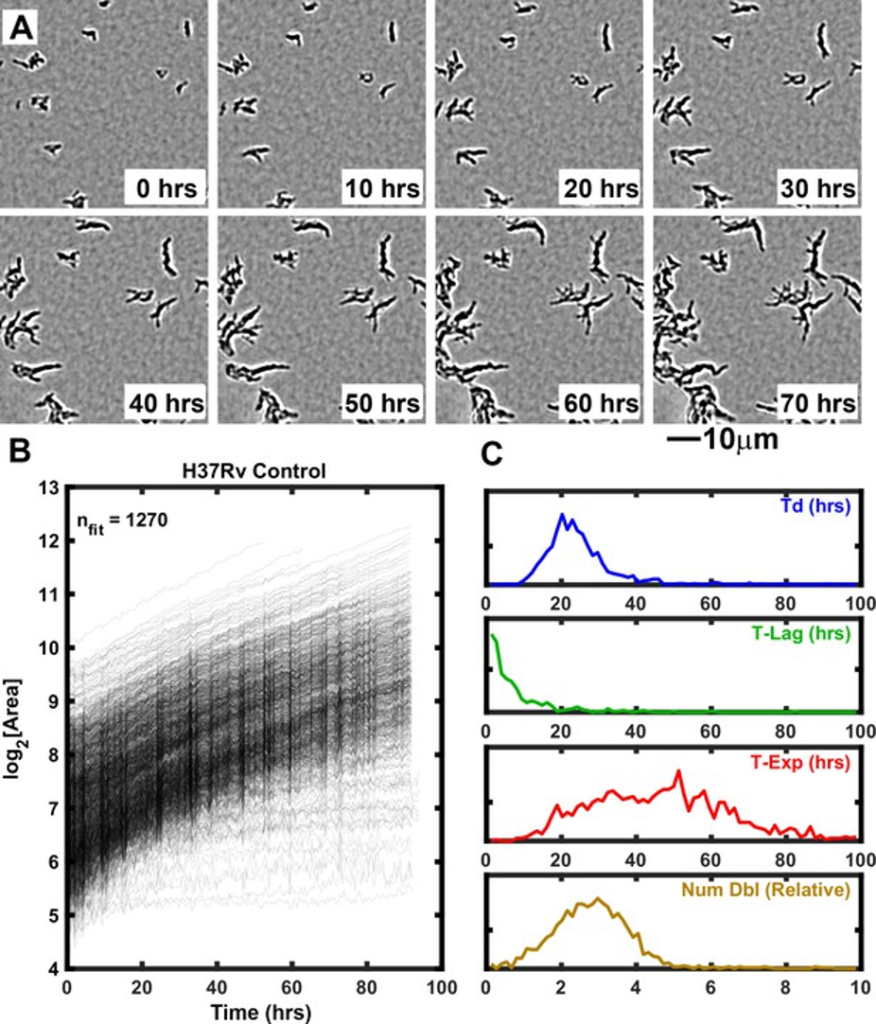
Growth of Mtb strain H37Rv into microcolonies over time observed with ODELAM Selected time-lapse snapshots of Mtb colonies growing on a solid medium. Mtb CFUs are shown growing on a 60 μm x 60 μm region of 7H9-GO agar over 96 hrs time course. (B) Growth curves for a total of 1276 colonies were recorded from a 1.2 mm x 0.9 mm region over 96 hrs and plotted. (C) Population histograms of the all extracted growth parameters (Doubling Time, Lag Time, Exponential Time and Number of doublings).
Antibiotics are required to treat a wide variety of microbial infections. However, with continued use of antibiotics, pathogens have developed antibiotic resistance that has rendered some antibiotics ineffective. Mtb, the microbe that causes tuberculosis, is particularly deadly, killing approximately 1.1 million people per year. Like other bacteria, Mtb has evolved mechanisms to resist the effects antibiotics. Understanding how antibiotic resistance emerges is critical for the development of countermeasures to battle emerging antibiotic resistance. We developed a technique named One-Cell Doubling Evaluation of Living Arrays of Mycobacterium, or ODELAM, that uses a microscope to automatically watch tens of thousands of individual Mtb cells grow over time as they are exposed to antibiotics. Remarkably, although cells are genetically identical, individual cells in a population respond differently to antibiotic drugs. This so-called antibiotic-induced population heterogeneity is key to how organisms like Mtb develop antibiotic resistance. The detailed growth measurements that ODELAM provides offer insight into the mechanisms of how antibiotics kill Mtb and what strategies Mtb develops to become resistant to antibiotics. The sensitivity of ODELAM allows us to detect different resistance states in individual clinical isolates. This is critical for detecting very small populations of resistance that can persist despite drug treatment. We anticipate these investigations will provide additional information for the design of new antibiotics and inform more effective application of antibiotics to patients.
RockEDU profiles NCDIR scientist Dr. Natalia Ketaren!
RockEDU Science Outreach at The Rockefeller University has featured NCDIR scientist Dr. Natalia Ketaren as their February #SCIONY (Scientists of New York)!
Read Natalia’s interview for RockEDU in the link below!

Research Spotlight – Structural basis of substrate recognition by a polypeptide processing and secretion transporter

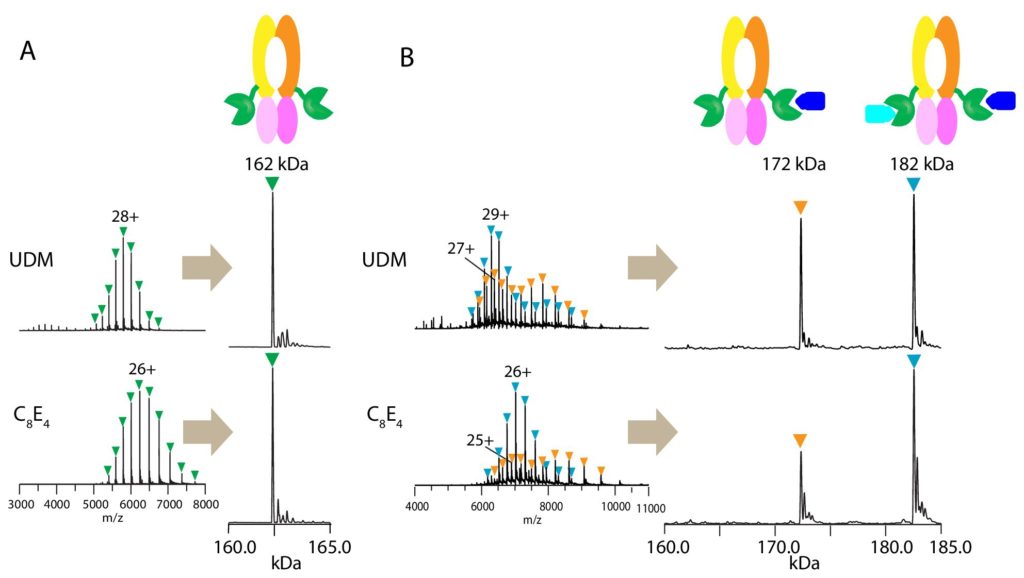
NCDIR scientists at The Rockefeller University: Dr. Dom Olinares (left) and Prof. Brian Chait, assisted fellow Rockefeller scientists in the lab of Dr. Jue Chen by using Native mass spectrometry to aid structural biology in determining the structure of a substrate-bound peptidase-containing transporter!
Peptidase-containing ATP-binding cassette transporters (PCATs) are a unique class of transmembrane transporters that secrete antimicrobial or quorum-sensing peptides. PCATs contain peptidase domains fused to the transmembrane transporter segments that cleave the polypeptide substrate prior to translocation.
Native mass spectrometry (MS) enables direct mass measurement of intact protein assemblies providing critical information on the composition and stoichiometry of protein complexes for integrative structural biology studies. NCDIR scientist Dom Olinares from the Chait Lab used native MS to determine the assembly state of and the number of bound substrates in the PCAT1 complex. The resulting native MS information greatly helped in the structural analysis of the transporter using cryo-electron microscopy by the laboratory of Jue Chen at The Rockefeller University.
Native MS analysis revealed that up to two substrates can bind the homodimeric PCAT1. The high resolution, cryo-EM structure of PCAT1 in complex with its substrate showed that two substrates are bound to the transporter, yet only one is positioned for cleavage and translocation. Overall, the study yielded insights on how substrate cleavage, ATP hydrolysis, and substrate translocation are coordinated in a PCAT transport cycle.
For more information, you can access the paper here: https://elifesciences.org/articles/51492