A landmark structural biology paper on understanding how a transcription-coupled repair protein disassembles a stalled transcription complex was recently published from the lab of Seth Darst and Elizabeth Campbell with the help of NCDIR scientist Dom Olinares from Brian Chait’s Lab.
Transcription is an essential and multi-step process wherein genetic information encoded in DNA is transcribed into RNA by the RNA polymerase (RNAP) complex. When the bacterial transcription elongation complex (EC) gets stuck on damaged DNA, an ATP-powered, transcription repair protein known as Mfd binds to the EC and displaces the stalled RNAP complex from the DNA.
To capture the structural details involved in the activity of Mfd, the group used single-particle cryo-EM. However, they initially encountered issues with sample preparation—the starting conditions used did not yield any Mfd-bound transcription complexes.
“We performed a sample quality check using the native mass spectrometry (MS) platform that was developed here at NCDIR and we mainly did not observe Mfd bound to the EC. So either Mfd was not binding at all or it quickly falls off after binding to RNAP. These are not optimal for cryo-EM analysis. To efficiently determine conditions that yield intact Mfd-bound complexes, we screened for samples using the same native MS platform by varying incubation times, DNA/RNA sequences, and Mfd preparation. It turns out that all these parameters were critical,” according to Dom.
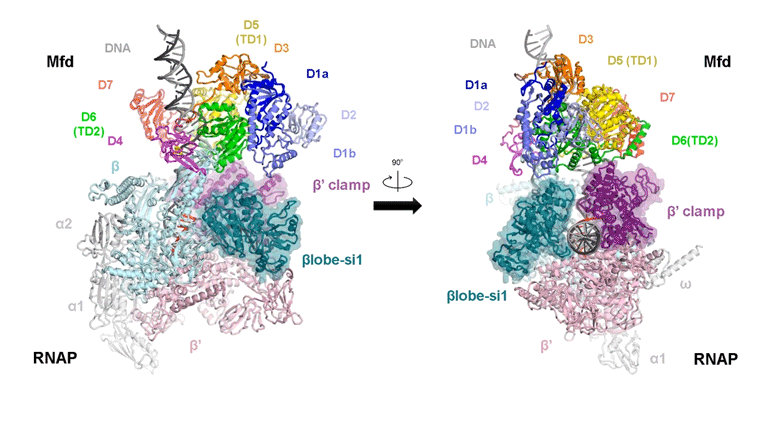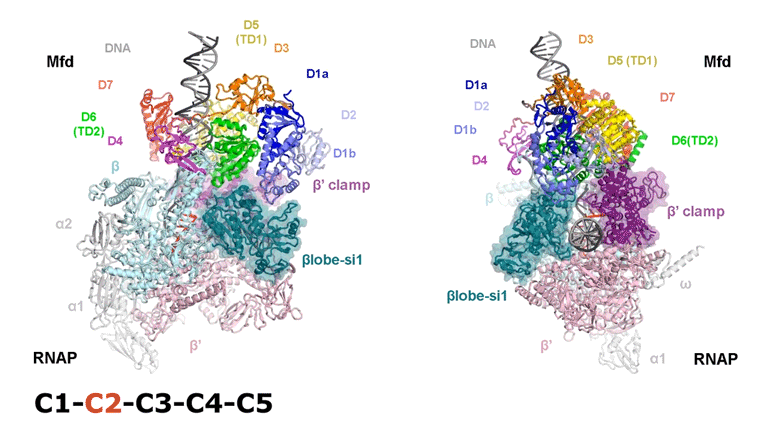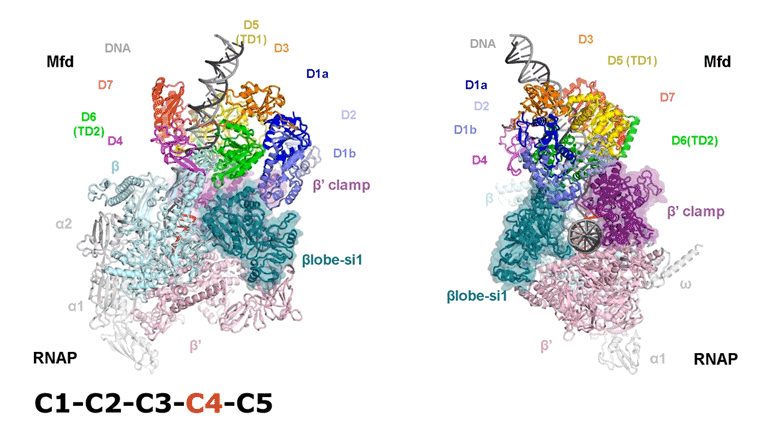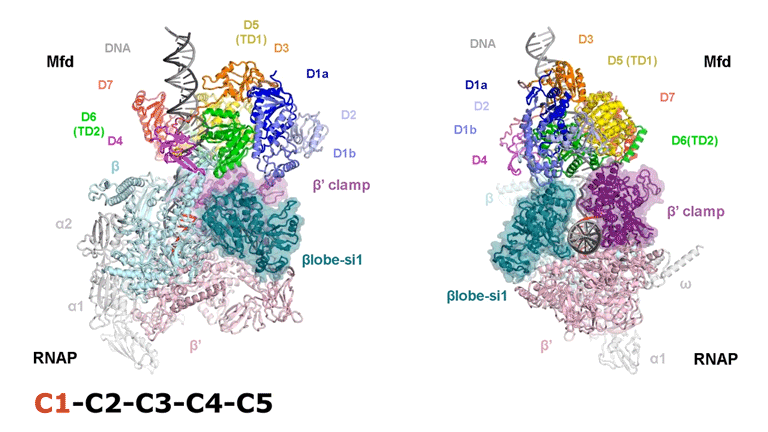
The video shows two orthogonal views of the Mfd-bound complex and cycles through the five structures of the Mfd-nucleotide hydrolysis (C1 → C2 → C3 → C4 → C5). At C2 → C3, Mfd opens the RNAP clamp and at C4 → C5, Mfd twists the βlobe-Si1 domain. The multiple domains of Mfd are colored and indicated accordingly (Kang et. al., 2021).
The native MS-based screening generated the best combination of sample conditions for preparing intact and homogeneous Mfd-bound transcription complexes for cryo-EM analysis. The result? Seven distinct structures that capture how Mfd engages and disassembles the transcription complex at different ADP/ATP states. Overall, the structural intermediates delineate how the initial interaction of Mfd with the stalled complex triggers a stepwise series of dynamic conformational changes, culminating in the stable engagement of Mfd with the EC and then ATP-powered disassembly of the stalled complex (see animation).
The paper is freely accessible here:
https://elifesciences.org/articles/62117
Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, Darst SA. “Structural basis for transcription complex disruption by the Mfd translocase.” Elife. 2021 Jan 22;10:e62117. PMID: 33480355.
