Integrative Modeling Platform
Welcome to the Integrative Modeling Platform (IMP)!
The goal of the Integrative Modeling Platform (IMP) is to contribute to a comprehensive structural characterization of biomolecules ranging in size and complexity from small peptides to large macromolecular assemblies. Detailed structural characterization of assemblies is generally impossible by any single existing experimental or computational method. This barrier can be overcome by hybrid approaches that integrate data from diverse biochemical and biophysical experiments (eg, x-ray crystallography, NMR spectroscopy, electron microscopy, immuno-electron microscopy, footprinting, chemical cross-linking, FRET spectroscopy, small angle X-ray scattering, immunoprecipitation, genetic interactions, etc…).
We formulate the hybrid approach to structure determination as an optimization problem, the solution of which requires three main components:
- the representation of the assembly,
- the scoring function and
- the optimization method.
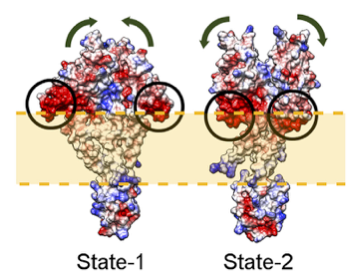
Figure 1: Multi-state modeling of the PhoQ two-component system
The ensemble of solutions to the optimization problem embodies the most accurate structural characterization given the available information.
We created IMP to make it easier to implement such an integrative approach to structural and dynamics problems. IMP is designed to allow mixing and matching of existing modeling components as well as easy addition of new functionality.
IMP provides an open source C++ and Python toolbox for solving complex modeling problems, and a number of applications for tackling some common problems in a user-friendly way. IMP can also be used from the Chimera molecular modeling system, or via one of several web applications.
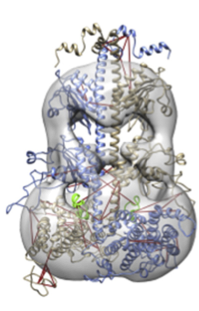
Figure 2: Model of the phosphodiesterase (PDE6)
IMP’s open source software is mostly available under the terms of the GNU Lesser General Public License (LGPL). (Some IMP modules are available under the GNU GPL instead.)
IMP is developed by Andrej Sali’s lab. To download, click here! Please contact us if you would like to learn more or request training in applying IMP to your research, please contact us here!
We encourage and support contributions from other laboratories through the IMP Community.
Additional downloads relating to IMP:
- Open-source version of the latest IMP code used to generate models for the spindle pole body core (above manuscript) is at https://github.com/salilab/imp(module spb) Models of the SPB core are available at https://doi.org/10.5281/zenodo.2603180
- Benchmark data for the sampling exhaustiveness test is at https://salilab.org/sampconOpen-source version of the IMP code used for optimizing representations is at https://github.com/salilab/optrep (module optrep).
- Benchmark data for the method to optimize coarse-grained representations for a system is at https://github.com/salilab/optimal_representation
- Open-source version of the latest IMP code used to generate models for the interaction of Spc110 with the gamma tubulin subcomplex are available at https://doi.org/10.5281/zenodo.4590125
