Nanobodies
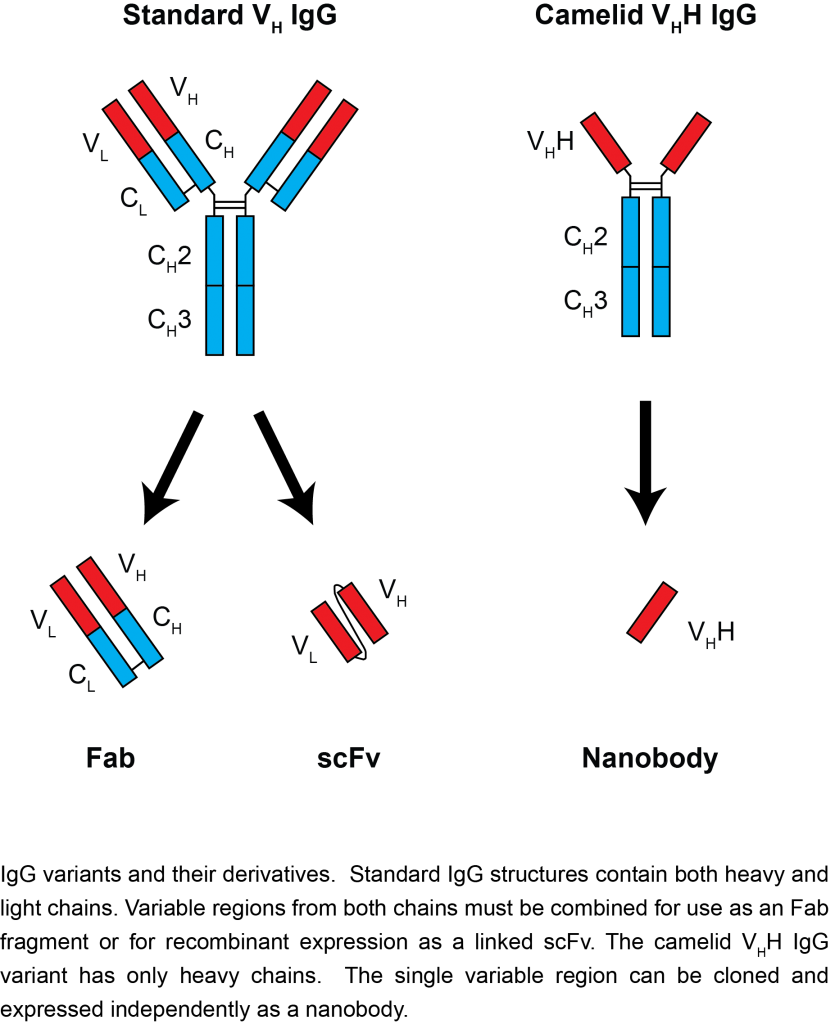
Nanobodies are a unique kind of monoclonal antibody derived from a camelid IgG variant, consisting of a single heavy-chain variable domain that can bind its antigen as strongly as a standard antibody. As nanobodies lack a light chain, they are both significantly smaller than standard antibodies, and have unique flexibility at their antigen-binding interface. This combination allows nanobodies to bind in different modes than typical antibodies, covering more chemical space and allowing binding to epitopes otherwise inaccessible to antibodies. Nanobodies are also significantly smaller (~15 kDa) and more stable than standard antibodies, and can be easily genetically engineered for additional functionality. With these advantages in mind, we and many others are actively pursuing nanobodies as improved reagents for multiple applications: antibody therapy and diagnostics, affinity isolations, imaging, and more.
“…nanobodies lack a light chain, they are significantly smaller than standard antibodies…”
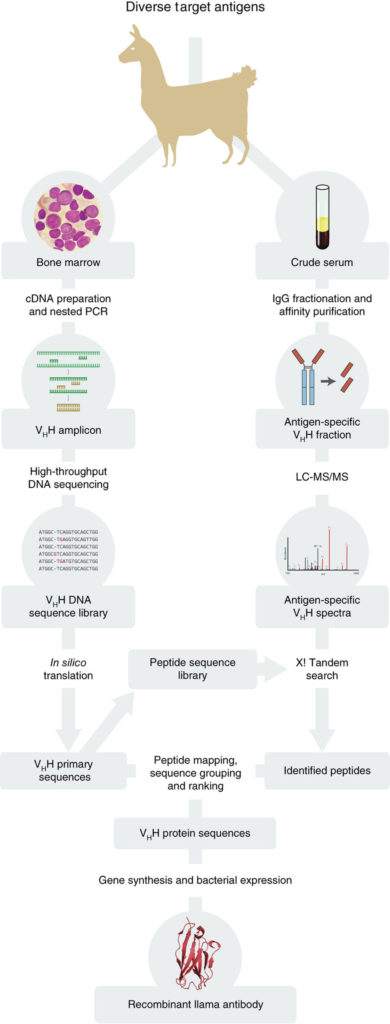
We have developed and optimized a new technology, termed Nanobody MiSeq/MS, which combines deep sequencing with mass spectrometry to rapidly generate large numbers of bacterially-expressed nanobodies with extremely high affinity and specificity. In our nanobody development pipeline, we first immunize llamas with the antigen target. After a strong immune response is elicited, we collect lymphocytes from bone marrow, highly enriched for active B cells. We then purify cDNA from these cells, and perform high-throughput sequencing on the PCR-amplified variable region (VHH) of heavy-chain-only IgG variants (HCAbs). In parallel, we collect animal sera, and affinity purify HCAbs with strong affinity and specificity to each antigen. The purified HCAbs are then analyzed by MS, and by correlating peptides to the DNA database of full-length sequences, we can identify candidate nanobody clones.
Llama-Magic is the automated pipeline that was used for identification of the Nanobody sequences and can be accessed here – Llama-Magic_GitHub. Llama-Magic allows upload of FASTA files containing reads from High-throughput DNA sequencing. Once uploaded, the reads will be automatically translated and digested to create an MS searchable database of tryptic peptides. Next, the MS (mgf) files can be uploaded for a selected tryptic peptide sequence database, and the parent and fragment error can be chosen for the X! Tandem search. Once the mgf files are uploaded, the X! Tandem search will be executed and the matching peptides saved. Then (1) annotation of CDR regions, (2) mapping of the identified peptides and (3) ranking and grouping of candidates are performed automatically, producing an interactive display of the candidate list showing detailed information regarding each sequence and its corresponding rank. Llama-Magic is implemented in Perl, HTML and JavaScript.
We first demonstrated the effectiveness of this approach by generating more than 30 high affinity nanobodies against GFP and mCherry.
“We freely provide these clones to the public, and have shared ~400 plasmids with more than 120 outside groups.”
Since this initial proof of principle, we have applied our nanobody generation pipeline to over 30 new antigens, generating a total of more than 150 unique nanobodies. Among our current interests, we are developing new nanobodies for use as therapeutics against cancer targets.
If you would like to learn more about our nanobody production pipeline, read it here! Also, please contact us if you are interested in a particular reagent/software or collaborating with us.
Latest NCDIR publication related to nanobodies:
Mast FD, Fridy PC, Ketaren NE, Wang J, Jacobs EY, Olivier JP, Sanyal T, Molloy KR, Schmidt F, Rutkowska M, Weisblum Y, Rich LM, Vanderwall ER, Dambrauskas N, Vigdorovich V, Keegan S, Jiler JB, Stein ME, Olinares PDB, Herlands L, Hatziioannou T, Sather DN, Debley JS, Fenyö D, Sali A, Bieniasz PD, Aitchison JD, Chait BT, Rout MP. “Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape.” Elife. 2021 Dec 7;10:e73027. doi: 10.7554/eLife.73027. PMC8651292